Effrina Yanti Hamid, S.T., M.T., Ph.D.
Pelaksana | Sekolah Teknik Elektro dan Informatika
Abstract
Myocardial infarction accounts for 49.2% of cardiovascular disease deaths, and the cTnI protein is a crucial target in diagnosing myocardial infarction. A peptide-based bioreceptor design using a computational approach is a good candidate to be developed for a rapid, effective, and selective detection method for cTnI although it is still lacking in study. Hence, to address the scientific gap, we develop a new candidate peptide for the cTnI biosensor by bioinformatics method and present new computational approaches. The sequential point mutations were made to the selected peptide to increase its stability and affinity for cTnI. Next, molecular docking was performed to select the mutated peptide, and one of the best results was subjected to the molecular dynamics simulation. Finally, the results showed that the best peptide showed the lowest affinity and good stability among other mutated peptide designs for interacting with the cTnI protein. In addition, the peptide has been tested to have a higher specificity towards cTnI than its major isomer, sTnI, through molecular docking and molecular dynamics simulation.
Keyword: cTnI, peptide-based biosensor, bioinformatics
Introduction
There is global burden of cardiovascular diseases (CVD), particularly myocardial infarction (MI), which is a leading cause of death. MI occurs when the heart muscle receives insufficient blood supply, necessitating early diagnosis to mitigate the health system’s load. Cardiac troponins, especially troponin I (cTnI), are highlighted as specific biomarkers for diagnosing MI. Although current detection methods like electrochemiluminescence are effective, they are costly and require specialized laboratories. Thus, there’s a need for more accessible, efficient diagnostic tools.
To address these challenges, researchers have explored biosensors utilizing biorecognition elements (BREs) like peptides. Peptides are attractive due to their low cost, stability, and specificity. Prior studies have demonstrated the potential of computational approaches for designing peptide-based BREs. This paper introduces a novel computational method for designing peptides targeting cTnI, optimizing their stability and specificity, and thereby providing a promising candidate for MI biosensors. We design some new peptides by selecting their interaction with the cTnI protein from the protein database and then optimizing the selected peptide design using the sequential point mutation. Hence, our study reveals the new potential candidate peptide as the cTnI protein biosensor also provides a new computational approach that can aid the optimization of the development of cTnI peptide-based biosensors.
Research Mode
The development of a peptide-based biosensor for cardiac troponin I (cTnI) was conducted using computational approaches. First, the cTnI protein structure was refined from its complex form in the RCSB database to create a standalone model. Binding sites were identified using the FTsite web server and literature analysis, enabling the selection of peptide residues within a 4 Å interaction radius. Peptide candidates were designed using trRosetta and validated for structural quality through ERRAT and Ramachandran plot assessments (Fig. 1). Molecular docking was performed via ClusPro and HADDOCK servers to evaluate binding interactions, with peptide B identified as the most promising candidate based on its low binding energy and strong hydrogen and salt bridge interactions.
To further optimize peptide B, sequential point mutations were conducted using the BeAtMuSiC web server to enhance binding affinity and interaction stability. The top three mutated peptides, alongside a wet-lab reference peptide, were analyzed using molecular dynamics simulations in GROMACS. Parameters such as Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Solvent Accessible Surface Area (SASA), and hydrogen bonding were evaluated over 25 ns. The best peptide (System 3) demonstrated strong and stable interaction with cTnI, outperforming both other candidates and the reference peptide. Selectivity tests confirmed that System 3 interacted more specifically with cTnI than with the similar skeletal troponin I (sTnI), reinforcing its suitability for biosensor applications.

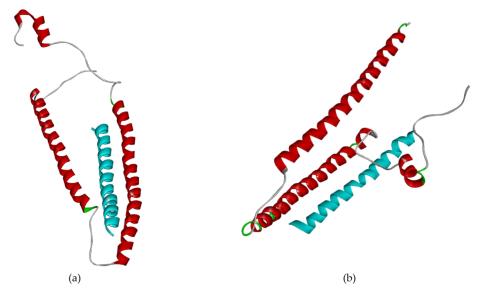
Discussion & Result
Initially, the structure of cTnI was modeled and refined using bioinformatics tools, ensuring high quality through validation metrics like ERRAT scores and Ramachandran plots. Four peptide sequences were developed, with one taken from literature as a control. These peptides were assessed for their structural properties and potential interactions with cTnI.
Through molecular docking, Peptide B was identified as the best candidate due to its low binding energy and stable interactions with cTnI, as confirmed by ClusPro and HADDOCK scores. Further analysis of hydrogen and salt bridge interactions revealed that residues LYS72, ARG79, GLU115, and THR129 in cTnI were consistently involved in binding, highlighting their importance in stabilizing the peptide-protein interaction within hydrophobic pockets. Peptide B underwent point mutations to optimize its binding affinity and interaction stability. Sequential mutation led to the generation of three improved peptides. Molecular dynamics simulations revealed that the peptide identified as System 3 demonstrated the best stability, compactness, and hydrogen bond interactions compared to other candidates and the literature peptide. It also showed minimal fluctuations, confirming its robust binding with cTnI.
Fig. 2 highlights the differential interaction poses of System 3 with cTnI and its major isomer, skeletal troponin I (sTnI). System 3 is stable in building hydrogen bonds during molecular dynamic simulation 25 ns. In contrast, System 5 failed to keep a con- stant interaction of hydrogen bonds. The peptide showed significantly stronger and more stable interactions with cTnI than with sTnI, supporting its specificity. It may be a cause of making System 5 not as stable as Sys- tem 3 according to the molecular dynamic result because hydrogen bond plays a vital role in the protein complex stabilization. This result establishes System 3 as a promising biosensor candidate for myocardial infarction diagnosis.
Conclusion
To summarize our result, we have successfully developed a cTnI protein-based biosensor using a computational approach. Peptide B with the sequence,
“WQTIYNLWAEKFDLQEKFKQQKYEINVERNRINDN”
becomes the best peptide that has the most stable and strong interaction with the cTnI protein. Furthermore, according to the selective test, peptide B is more stable and stronger for the cTnI protein compared to the sTnI protein. Therefore, our study provides a new potential candidate for myocardial infarction diagnosis to ease the burden of cardiovascular disease.